
NEXTFLEX UDI-UMI Barcodes (1-48)



| Feature | Specification |
|---|---|
| Automation Compatible | Yes |
| Product Group | Barcodes |



Supporting products you might need
Product information
Overview
- Full length 10 base-pair UDI + 9 base-pair UMI adapters supplied in a 96 well plate for direct use manually or automated liquid handlers.
- Reduces index hopping on Illumina and Element sequencers.
- UMI enabled consensus building supports accurate duplicate removal and variant allele detection down to 0.1 % in ctDNA and low input RNA seq datasets.
- Compatible across whole genome, targeted DNA seq, and mRNA seq workflows including those with PCR-free protocols
- Drop in compatibility with TruSeq® style library prep chemistries from Revvity, Watchmaker, Illumina®, NEB, KAPA, and Twist, no protocol changes required.
- Supports 8–96 multiplexing per lane
- Every lot is functionally verified and tested for index purity (>99.9%) by sequencing.
Additional product information
Integrated UD + UMI architecture
Each kit arrives as a 96 well plate, every well holding a full length adapter pair with a 10 bp Unique Dual Index (UDI) and a 9 bp Unique Molecular Identifier (UMI). The barcode is compatible with standard TruSeq® workflows. Placing distinct 10 base UDI on both ends of every fragment reduces misassignment and index hopping rates below 0.1-0.3 % on Illumina® and Element® flow cells.

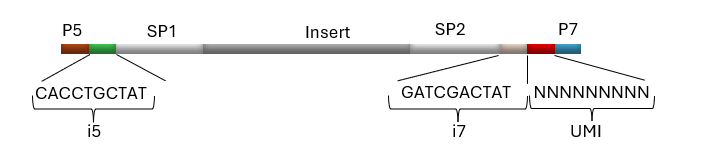
Figure 1. Example of a library containing NEXTFLEX UDI-UMI Adapters, where Insert = DNA or RNA fragment from a sample, P5 and P7 = Flow cell binding sites for Illumina® platforms, SP1 and SP2 = Binding sites for sequencing primers, i5 and i7 = Short pair of 10 bp sequences used to identify a particular sample (UDI), and UMI = 9 bp sequence used to uniquely tag each molecule within a library.
The 9 bp UMI, also known as a molecular barcode, is attached to each original DNA or RNA fragment prior to amplification. During analysis, reads that share the same UMI and map to identical genomic coordinates are grouped together. These groups are collapsed into a consensus sequence, removing PCR duplicates and correcting random sequencing errors. If you’re setting up your analysis pipeline, review the common bioinformatic challenges with UDI-UMI data we outline here. Because every starting molecule carries a distinct tag, counting unique UMIs after deduplication provides a direct measure of library complexity (i.e., how many unique molecules were captured versus how many were merely re sequenced). This molecular level view supports confident detection of low frequency variants down to 0.1% in ctDNA and low input projects, while ensuring that sequencing depth is translated into true biological coverage rather than inflated duplicate counts.
Each UMI is a 9 nt sequence drawn from a roughly equimolar mix of A, T, C, G bases yielding 4⁹ ≈ 262 k unique tags. The large number of tags keeps UMI-collision rates low, even in high depth runs, while the uniform base composition also improves cluster calling and phasing on Illumina® and Element® flow cells, ensuring that UMI information is read accurately for downstream duplicate removal and variant calling.
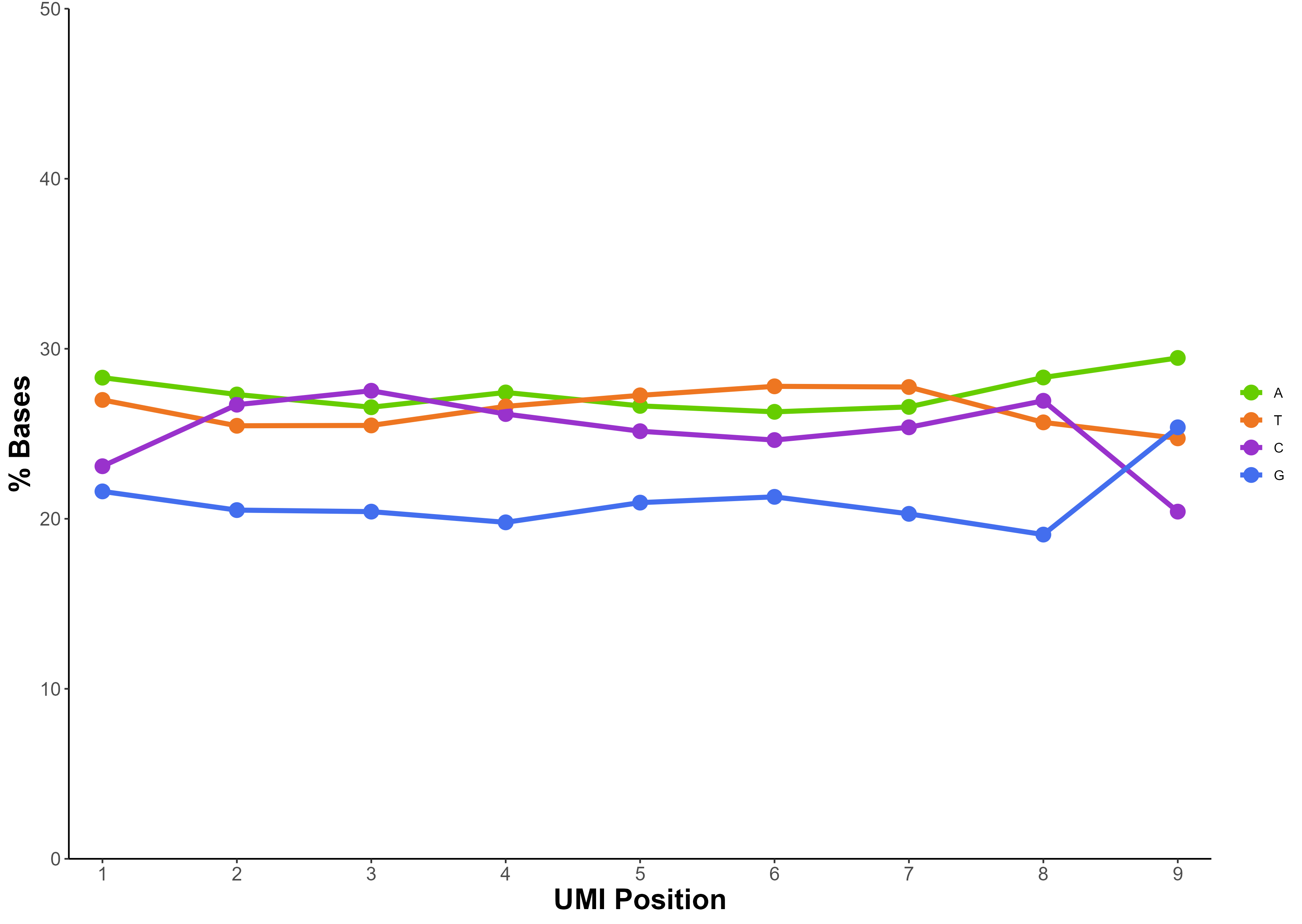
Figure 2. Balanced nucleotide composition across the 9 bp Unique Molecular Identifier (UMI) in 24 NEXTFLEX UDI UMI libraries prepared from 0.1–1000 ng human gDNA with the NEXTFLEX Rapid XP v2 DNA seq Kit and sequenced on an Illumina® MiSeq™ system, demonstrating high UMI diversity and stable library complexity from low to high input DNA.
Quantitative sequencing
When starting from low input, FFPE or GC skewed samples, extra PCR cycles can magnify polymerase bias and inflate duplicate counts, making alignment based deduplication unreliable, especially for highly expressed transcripts. By reducing overrepresentation from PCR, the NEXTFLEX UDI-UMI adapters improve the linear correlation between true and observed expression.
Rare variant analysis
Ultra low frequency variants (down to 0.1 % VAF) sit at the noise floor of conventional sequencing. NEXTFLEX UDI-UMI consensus reads strip away random polymerase and sequencing errors, while PCR free prep eliminates the polymerase step altogether, preventing artefacts that can masquerade as real variants. The combination delivers confident rare variant calls in ctDNA, targeted panels, low input RNA samples, and heterogeneous tumor biopsies even at the deep coverages typical of these sample types. See our deep dive on the power of UMIs in cancer research for data examples
Specifications
| Automation Compatible |
Yes
|
|---|---|
| Barcodes |
1 - 48
|
| Product Group |
Barcodes
|
| Shipping Conditions |
Shipped in Dry Ice
|
| Unit Size |
48 rxns
|
FAQs
-
How many unique molecular tags are available with the bp UMI?
-
What sequencing depth is required to detect 0.1 % variant allele frequency (VAF) with simplex UMIs?
-
Do I need duplex UMIs if I want to go below 0.1 % VAF?
-
Can these adapters be used in PCR free workflows?
-
How do I collapse UMI families and remove PCR duplicates?
-
Are the adapters automation friendly for 96 well liquid handlers?
-
What are the recommended storage conditions and shelf life for NEXTFLEX UDI UMI adapter plates?
Resources
Are you looking for resources, click on the resource type to explore further.
Considerations when selecting the right barcoded adapters
This document provides guidance on the applications in which NEXTFLEX UDI-UMI Barcodes can be used.
This flyer illustrates the breadth of the NEXTFLEX RNA-seq Portfolio
The need for better profiling is clear, and technological advances are making it possible to optimize clinical trials and...
Related reagents


Loading...
How can we help you?
We are here to answer your questions.






























