
Automated homogenization and DNA extraction from murine GI tract tissue, cultures, and mock community for 16S sequencing
The human microbiome is a complex ecosystem of microbial organisms and its role in maintaining physiological functions such as digestion, immunity, and cognition is recognized, fueling much of its research for an even better understanding. A variety of sample types, including stools, swabs, and tissue are used, along with different lab-to-lab sample prep methods. Within the microbiome community, different organisms have different susceptibility for lysis generally, making consistent and complete lysis difficult.
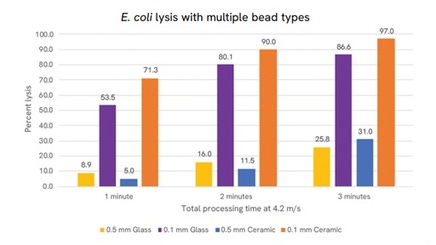
This application note explores bead-beating as a robust technology for physical lysis of microbes from mouse GI tissue into automated extraction. A variety of bead mixes were also used to optimize lysis of a variety of different organisms to evaluate lysis efficiency. Last, extraction bias from this methodology was evaluated using a mock community standard. Lysates were carried through to automated extraction of the chemagic 360 nucleic acid extractor and NEXTFLEX 16S sequencing kits.
For research use only. Not for use in diagnostic procedures.
To view the full content please answer a few questions
Download Resource
Automated homogenization and DNA extraction from murine GI tract tissue, cultures, and mock community for 16S sequencing




























