Explore our solutions
Customer experiences
Automated DNA extraction methods are revolutionizing genetic research, as demonstrated by these two insightful webinars. Karine Auribault from Paris and Emily Farrow from Kansas will share their experiences implementing high-throughput DNA isolation techniques in hospital settings. These presentations underscore the critical importance of efficient (HMW) DNA extraction in enabling a wide range of genetic tests and assays, from MLPA and Sanger sequencing to cutting-edge long read sequencing.
Suggested links:

Webinar review: High quality DNA for genetic testing

Unleashing the power of genomics: A webinar review
HMW DNA extraction for long read sequencing
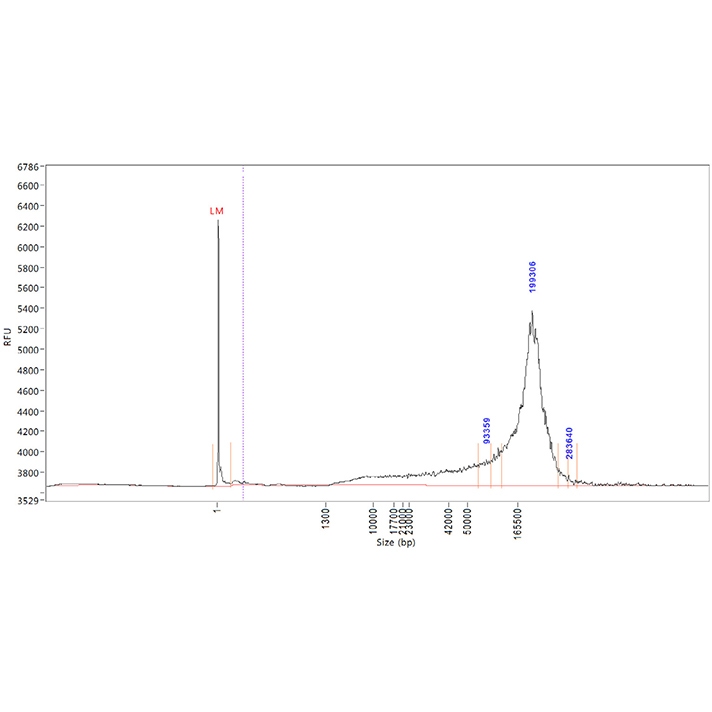
Learn more about efficient HMW DNA isolation in our Application Note. We compared extraction performance from blood based on DNA yield, purity, and integrity between two automated nucleic acid extraction systems.
Suggested links:
Comparison of automated nucleic acid purification systems HMW DNA extraction efficiency
Biobanking
Drawing on extensive expertise in high-throughput extractions across diverse sample types such as blood, buffy coat, swabs, and saliva, we have established a global presence in biobanking. Our chemagic nucleic acid isolation ensures archive-quality specimens, and our ability to automate entire workflows, from primary sample handling to downstream QC and assay setup, has positioned us as a trusted collaborator in various population genetic cohort studies.
Our contributions extend to notable initiatives like the "All of Us Research Programme" from the Mayo Clinic7, the German National Cohort/NAKO Gesundheitsstudie8, and the British Initiative, including the 100,000 Genomes Project, among others.

Suggested links:
Dried blood spots for advanced DNA sequencing
Automated nucleic acid isolation fulfilling biobanking needs

High quality DNA isolation suitable for ultra rapid sequencing
Featured products


References:
- Lang K, Wagner I, Schöne B, et al. ABO allele-level frequency estimation based on population-scale genotyping by next generation sequencing. BMC Genomics. 2016;17:374. Published 2016 May 20.
- Beyter D, Ingimundardottir H, Oddsson A, et al. Long-read sequencing of 3,622 Icelanders provides insight into the role of structural variants in human diseases and other traits. Nature Genetics. 2021 Jun;53(6):779-786.
- Steiert TA, Fuß J, Juzenas S, et al. High-throughput method for the hybridisation-based targeted enrichment of long genomic fragments for PacBio third-generation sequencing. NAR Genom Bioinform. 2022;4(3):lqac051. Published 2022 Jul 13.
- Schmidt J, Berghaus S, Blessing F, et al. Genotyping of familial Mediterranean fever gene (MEFV)-Single nucleotide polymorphism-Comparison of Nanopore with conventional Sanger sequencing. PLoS One. 2022;17(3):e0265622. Published 2022 Mar 17.
- Watson CM, Crinnion LA, Hewitt S, et al. Cas9-based enrichment and single-molecule sequencing for precise characterization of genomic duplications. Lab Invest. 2020;100(1):135-146.
- Watson CM, Crinnion LA, Simmonds J, Camm N, Adlard J, Bonthron DT. Long-read nanopore sequencing enables accurate confirmation of a recurrent PMS2 insertion-deletion variant located in a region of complex genomic architecture. Cancer Genet. 2021;256-257:122-126.
- Valentin N, Camilleri M, Carlson P, et al. Potential mechanisms of effects of serum-derived bovine immunoglobulin/protein isolate therapy in patients with diarrhea-predominant irritable bowel syndrome. Physiol Rep. 2017;5(5):e13170.
- Kousathanas A, Pairo-Castineira E, Rawlik K, et al. Whole-genome sequencing reveals host factors underlying critical COVID-19. Nature. 2022;607(7917):97-103.